2 R universe
To paraphrase, R is a dialect of another programming language, namely S. You can read more about the history of R (and S) here. Long story short, R is a programming language derived from S that was available only for commercial packages. R was created by Ross Ihaka and Robert Gentleman in 1991 at the University of Auckland, New Zealand. In 1995, it became an open source code thanks to contributions by Martin Mächler.
In this short book, I will use interchangeably r and R.
The online and free book by Roger D. Peng R programming for data science is a good further reading for those interested.
The online and free book by Oscar Baruffa Big book of R is an excelent collection of available resources to learn and master R.
R (the console and language)
When most people talk about r they mean both the programming language and a console. Unless they are IT experts who can make the distinction with ease. But, for the purpose of the seminar, or as a typical r-user for what is worth, it really doesn’t matter.
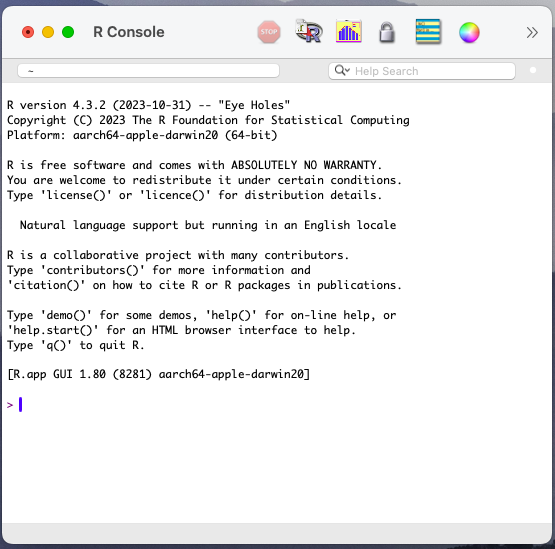
When working with r one needs a designated console for writing the code, and this is easy to detect as r-console (see Figure Figure 2.1).
To download R, go to the cran website and select the file suitable for your operating system. Unzip or install that file and the r-console will be installed on your machine.
The basics (the very basics!)
This seminar will cover only the very absolute basics of working in r. Designated courses are available at the university and elsewhere as part of summarschools or workshops. Of course, one can learn r using the freely available online content. Use YouTube and Google for that. For example, this online resource is a good starting point.
The first thing to notice in the r console is the symbol > followed by the text placer |. This specifies the line where to write the r code.
Once the code is written and the key Enter is pressed, the code basically runs or is computed by the machine which returns an outcome. (Here the symbol > is not visible but the outcome line can be identified through the use of squared brackets[…])
2+2[1] 42.0.1 Objects
It is useful to work with objects in r. That is, whatever code you write, place it into an object and then run the object itself. See below.
# no object created
2+2[1] 4# object is first created and then run
sum<-2+2
sum[1] 4Using objects simplifies a lot the work flow because you can combine objects in any way you can imagine!
# creates a second object called mean
mean<-mean(c(1,2,5,7,8,9))
mean[1] 5.333333# and then adds the two objects 'sum' and 'mean' together
result<-sum+mean
result[1] 9.3333332.0.2 Vectors
There are multiple types of objects that one can create in r. The most important ones are vectors and data tables.
For simplicity reasons, vectors can be numeric, character strings or logical. A vector is scalable meaning that it can hold up to a gazilion of elements.
# example of numeric vectors
vec1<-c(1,3,66,9,121)
vec1[1] 1 3 66 9 121# example of character string vector
vec2<-c("A","Ab","This or that","C","d")
vec2[1] "A" "Ab" "This or that" "C" "d" # example of logical vector
vec3<-c(TRUE,TRUE, FALSE, TRUE)
vec3[1] TRUE TRUE FALSE TRUEOne can do all sorts of things with and to vectors. See for example here.
2.0.3 Data tables
Data tables combine multiple vectors. Data tables can combine all sorts of vectors and can have varying internal structures. When one downloads (or uses one own’s) dataset, that is typically a data table in a specific format, .sav for SPSS or .xlsx for Microsoft Excell. Data formats can also be .dat, .csv, .asci and so on.
A data table in r comprises multiple vectors and involves an organization wherein typically rows represent entries in the data table and columns represent vectors of the data table. In other words, rows represent cases and columns represent variables.
# create a simple data table
df<-data.frame(col1=vec1,
col2=vec2)
df col1 col2
1 1 A
2 3 Ab
3 66 This or that
4 9 C
5 121 d# one can then access the varying elements of the data table
# access col1
df[,1][1] 1 3 66 9 121# access first row
df[1,] col1 col2
1 1 A# access entry at first row and col1
df[1,1][1] 1One can perform all sorts of actions on the data table as a whole or on elements of the data table.
# checks the elements of the data table
str(df)'data.frame': 5 obs. of 2 variables:
$ col1: num 1 3 66 9 121
$ col2: chr "A" "Ab" "This or that" "C" ...One can see that col1 is a numeric num vector and col2 is a character string char vector.
# provides a summary of the data table
summary(df) col1 col2
Min. : 1 Length:5
1st Qu.: 3 Class :character
Median : 9 Mode :character
Mean : 40
3rd Qu.: 66
Max. :121 One can see that different summary stats are available for num and chr vectors.
# performs an addition on the numeric vector of the data table
df[,1]+100[1] 101 103 166 109 221Functions
To be entirely honest, r functions are something a bit advanced. But, some rudimentary functions can be written by beginners too. The trick is to figure out what is repetitive in the code that one wants to write. This logic proves useful when one needs to apply a command on a number of objects for an undetermined number of times.
Functions are easy to spot in R because they are labeled as such and have a unique code structure: function(){}.
The rule of thumb is () defines the elements that are fed into the function while {} contains the function itself.
Here is an example. We use a dataset that comes pre-installed with R (iris), perform an addition on all the numerical variables and then write a function to simplify the task.
# see the first ten rows of the pre-installed dataset iris
head(iris) Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5.0 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosa# numerical columns are then columns 1 through 4
# adds 3 to all numerical columns
head(iris[,1:4] + 3) Sepal.Length Sepal.Width Petal.Length Petal.Width
1 8.1 6.5 4.4 3.2
2 7.9 6.0 4.4 3.2
3 7.7 6.2 4.3 3.2
4 7.6 6.1 4.5 3.2
5 8.0 6.6 4.4 3.2
6 8.4 6.9 4.7 3.4# add 77 to all numerical columns
head(iris[,1:4] + 77) Sepal.Length Sepal.Width Petal.Length Petal.Width
1 82.1 80.5 78.4 77.2
2 81.9 80.0 78.4 77.2
3 81.7 80.2 78.3 77.2
4 81.6 80.1 78.5 77.2
5 82.0 80.6 78.4 77.2
6 82.4 80.9 78.7 77.4# write a function
# this function takes two arguments: a dataset 'df' and a constant 'n'
func1<-function(df,n){
tmp <- Filter(is.numeric, df) # we first filter the dataframe for numeric columns
tmp + n # we then add the constant to all the numeric columns
}
# we apply the function and add 3 to all numeric columns of iris
# we only ask to see the first ten rows of the outcome using head()
head(func1(iris,3)) Sepal.Length Sepal.Width Petal.Length Petal.Width
1 8.1 6.5 4.4 3.2
2 7.9 6.0 4.4 3.2
3 7.7 6.2 4.3 3.2
4 7.6 6.1 4.5 3.2
5 8.0 6.6 4.4 3.2
6 8.4 6.9 4.7 3.4# we apply the function and add 99 to all numeric columns of another pre-installed dataset 'mtcars'
# we only ask to see the first ten rows of the outcome using head()
head(func1(mtcars,99)) mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 120.0 105 259 209 102.90 101.620 115.46 99 100 103 103
Mazda RX4 Wag 120.0 105 259 209 102.90 101.875 116.02 99 100 103 103
Datsun 710 121.8 103 207 192 102.85 101.320 117.61 100 100 103 100
Hornet 4 Drive 120.4 105 357 209 102.08 102.215 118.44 100 99 102 100
Hornet Sportabout 117.7 107 459 274 102.15 102.440 116.02 99 99 102 101
Valiant 117.1 105 324 204 101.76 102.460 119.22 100 99 102 100Packages
An R package contains code, documentation, and sometimes even data. These packages are developed to serve a specific purpose such as simplifying a work routine or perform advanced computational routines. Packages can be downloaded for free and then immediately used. Of course, everyone can write an R package, which of course is not a easy thing to do. But if at any point and for whatever reason you need to, then know that it is possible.
Everything one needs to know about packages can be found in this comprehensive book by Hadley Wickham1 and Jennifer Bryan.
r packages use the philosophy of working with functions to simplify otherwise highly complex code. Some of the fundamental packages to start with are tidyverse (for data preparation and manipulation but also contains several other useful packages like ggplot2 for creating graphics). Other packages that are the focus of this seminar are rmarkdown (the fundamentals of Chapter 3 through Chapter 5), quarto (needed for self-publishing books and website; covered in Chapter 4),tinytex (for latex distributions aka. creating PDFs), shiny (for web applications; covered in Chapter 5).
What you absolutely need to know about packages is that the vast majority do not come pre-installed with the r console but can be installed by request. Installing any package in R follows this basic routine:
# installs `tidyverse`
install.packages("tidyverse")
# makes it available for R on your local machine
# this step is crucial if you want to have access to all the containing function
library(tidyverse)One trick that I think it is absolutely simple to use but can save you a lot of nerves is using the package pacman to install any other packages. The nice thing about it is that pacman can first check if a package is already installed on the local machine and if not, it downloads it and installs it from Cran.
We can now install the basic packages needed for the seminar and mentioned above.
# first, we install the `pacman` package
install.packages("pacman")
# then, we use the function `p_load` from the `pacman` package to install `tidyverse`, `rmarkdown`, `shiny` packages
pacman::p_load(tidyverse,rmarkdown,bookdown,quarto,shiny)(Almost) Every package has a designated website. Visit the package website for examples on how to use and also to identify the functions contained. For example https://www.tidyverse.org/
Call the package documentation by typing in a question mark followed by the name of the package or function contained in a package. For example ?tidyverse
Let’s see as an example how the function filter from the universe of packages tidyverse works. Before that, I want to introduce the pipe operator %>%2 which is instrumental for r users. And it simplifies a lot the work flow!
%>% follows the logic of, simply and un-elegantly put, “work that happens in the background until the desired output is retrieved”. It also means that using %>% you can compress into one code otherwise a long chain of steps that involve creating objects which are then subjected to new operations.
# apply the function filter to the dataset mtcars
# we filter the column cyl such that only cars with a cyl < 5 are displayed
head(mtcars) mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1mtcars %>% filter(cyl < 5) mpg cyl disp hp drat wt qsec vs am gear carb
Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1
Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2
Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2
Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1
Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1
Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1
Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2
Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2
Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.60 1 1 4 2# we filter the column cyl such that only cars with a cyl exactly equal to 8 are displayed
mtcars %>% filter(cyl == 8) mpg cyl disp hp drat wt qsec vs am gear carb
Hornet Sportabout 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2
Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4
Merc 450SE 16.4 8 275.8 180 3.07 4.070 17.40 0 0 3 3
Merc 450SL 17.3 8 275.8 180 3.07 3.730 17.60 0 0 3 3
Merc 450SLC 15.2 8 275.8 180 3.07 3.780 18.00 0 0 3 3
Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0 3 4
Lincoln Continental 10.4 8 460.0 215 3.00 5.424 17.82 0 0 3 4
Chrysler Imperial 14.7 8 440.0 230 3.23 5.345 17.42 0 0 3 4
Dodge Challenger 15.5 8 318.0 150 2.76 3.520 16.87 0 0 3 2
AMC Javelin 15.2 8 304.0 150 3.15 3.435 17.30 0 0 3 2
Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0 3 4
Pontiac Firebird 19.2 8 400.0 175 3.08 3.845 17.05 0 0 3 2
Ford Pantera L 15.8 8 351.0 264 4.22 3.170 14.50 0 1 5 4
Maserati Bora 15.0 8 301.0 335 3.54 3.570 14.60 0 1 5 8As an example of the usefulness of the pipeline operator %>%, let us apply a double filter. First, on the column cyl and then on the column horse power hp.
# without the pipeline operator
a<-mtcars %>% filter(cyl < 5)
b<-a %>% filter(hp > 100)
b mpg cyl disp hp drat wt qsec vs am gear carb
Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.9 1 1 5 2
Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.6 1 1 4 2# with the pipeline operator
mtcars %>% filter(cyl < 5) %>% filter(hp > 100) mpg cyl disp hp drat wt qsec vs am gear carb
Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.9 1 1 5 2
Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.6 1 1 4 2Of course, this example is too simplistic but imagine having to write a gazillion of lines of code when you could reduce that to a couple. Throughout the seminar we use the pipe operator %>% almost everywhere!
Base R vs. Packages
A fair warning!
Base R is complex but stable. Packages are simple to use but depend on the community for their maintenance. So, the decision is to use something complex but stable or simple but unstable.
For the purpose of this seminar, and for most of the things a regular R-user needs, working with packages is indeed the way to go.
If at any point, you are concerned that the package(s) you use can get outdated, I recommend using the [sic!] package groundhog which ensures reproducible code. This package basically goes back in time and installs on the local machine the desired version of the package.
See how it works on this website.
RStudio
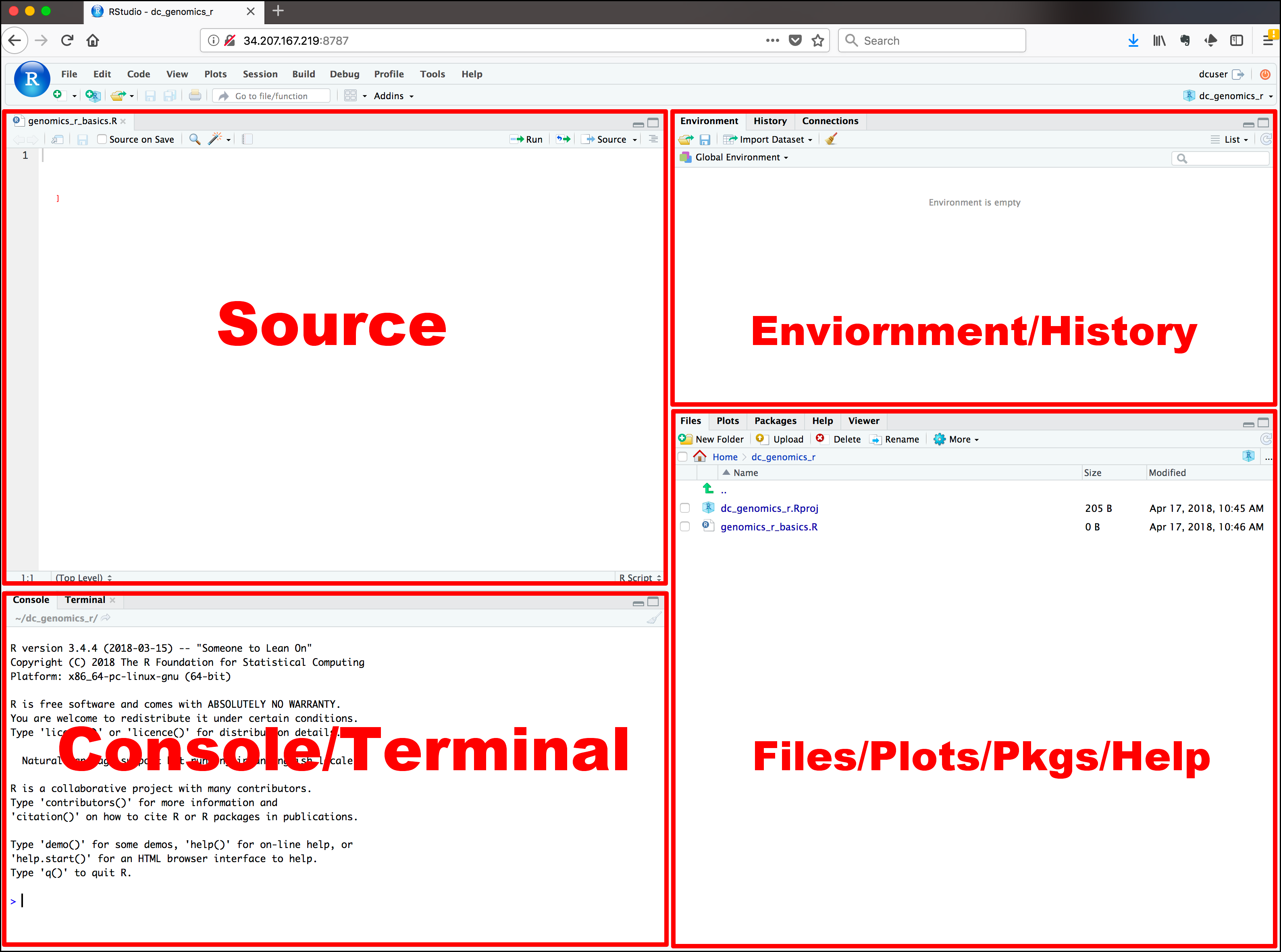
In Figure 2.3 you can see the four panels of RStudio, the (a) Console/terminal, (b) Source, (c) Environment/history, and (d) Files/plot/packages/help.
a) Console/terminal Here is where the
rconsole is integrated inRStudio. You can type in your code, have your results previewed, as well as any errors (those happen quite a lot) that occur in your coding.b) Source This panel is where we will do most of the work throughout the seminar. Think of this panel as the notebook – you write, you draw, you comment on your own work, etc. This panel allows you to communicate with the source material, which can be
r(the language),html(the language) and also lets you populate with content the files needed for the website, for instance.c) Environment/history This panel is a place where you can see the history of your work. It saves for you the code you ran (either in the console or source panels) and also it contains sort of short-cuts to any data-related work you might have done.
d) Files/plot/packages/help This panel allows you to preview what you’ve communicated to the machine (laptop) to do. You will note there are several tabs, but the most important one for the seminar are:
Filesis sort of Windows explorer in Windows or Finder on Mac OS. It is here that you can navigate between folders on the local machine, delete, rename, or more. Here you can also open files in the source panel.Packagesgives you an overview of packages that are installed and active on the local machine.Helpis, well, where you will see helpful information about a function or package.
On this youtube channel there is a helpful beginners guide on R and RStudio. Take some time to familiarize yourself with them.
If your RStudio version is 2024.04. or newer, you should note in the Environment/ History panel a tab “Turorial”. That panel contains tutorials for working in R. Install first the package learnr as indicated and let yourself guided through a number of interactive exercises.
Advanced resources
Together with a colleague, Dr. Ranjit SINGH from GESIS - Leibniz Institute for the Social Sciences, I prepared a workshop on r for beginners. All the material is open access via GitHub.
You can clone the repository on your local machine and do all the exercises.
Navigate first to the page of the repository and then clone it to your local machine: https://github.com/adrianvstanciu/rworkshop_open.
He is THE
rexpert. See his website.↩︎The pipe operator itself is introduced most comonly in the package
dplyrcontained in the universe of packagestidyverse. But, it can be used differently in other packages too.↩︎